Project Aims
Ordium is a bioinformatics analysis platform with built in sample tracking Lab Information Management System (LIMS) and Electronic Health Record (EHR) tools
Navigate this page with left and right arrow keys, or by clicking the boxes below
Lab Informatics System
Ordium LIMS/LIS (Lab Information Management System, or Lab Informatics System) tracks organisms and derived sample material, assists with sample prep, and tracks consumables in the lab, with support for modern assays and sequencing technologies. It provides a seamless architecture and workflow with downstream analyses.
Being web based, it requires little to no up-front investment and will most likely work on your existing hardware.
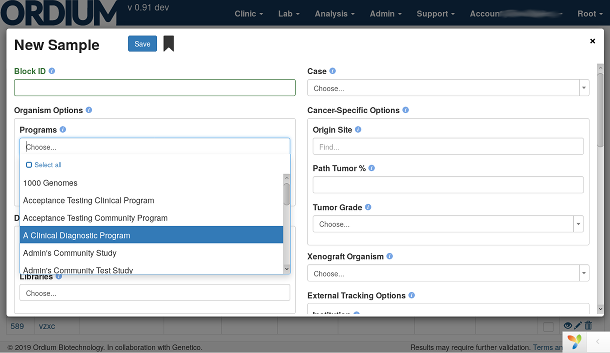
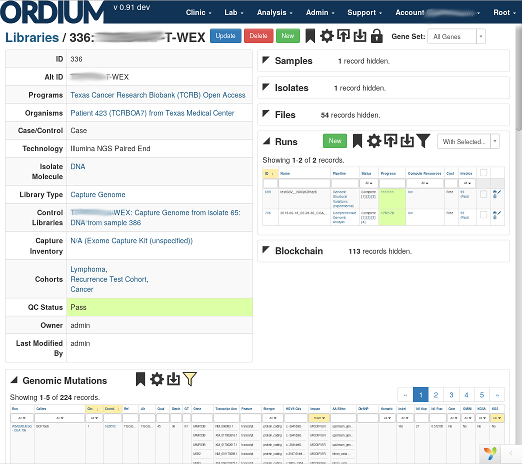
Relationships between biological organisms (such as patients), samples and aliquots of material from them, such as biopsies or culture cells, sequence libraries, consumable inventory such as chemistry and reagents, and more can be captured. The intuitive interface helps keep your data consistent and error-free.
Share data securely and restrict access to users within your institution, lab or other working group, using powerful access control and auditing features in our HIPAA-compliant environment.
Questions? Send Us a Message
Electronic Health Record System
Ordium provides EHR/EMR tools for clinicans and researchers. Although it's not intended to replace enterprise EHR systems for hospitals or large clinics, it does have many of the same features. It's simple and easy to use, and works seamlessly with new technologies such as Next-Gen Sequnencing (NGS).
Security and compliance built-in. Ordium runs in a HIPAA compliant environment, and uses an access control system that protects patient privacy, while allowing data sharing across organizational groups and institutions. It can restrict access to private data, and opt patients in to research studies and data sharing with the proper informed consent.
It's a simple yet powerful platform. Sign Up for Free
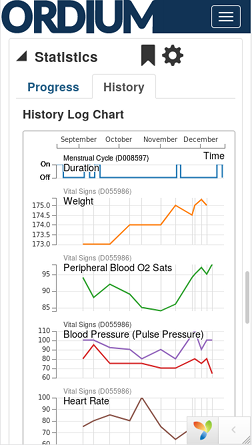
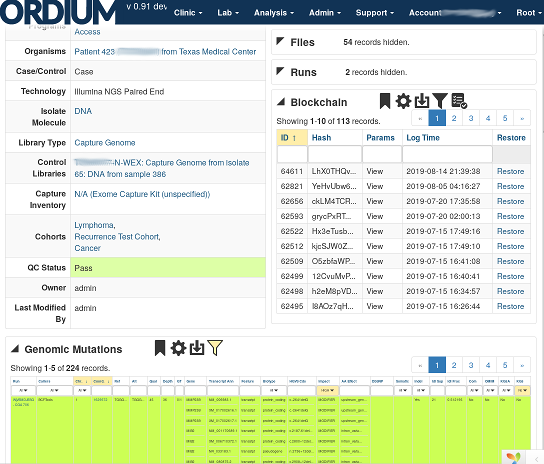
Enterprise spport is available. Choose either the free, community supported Basic Tier subscription, or a Prime Tier subscription, which includes access to voice and email tech support, and faster more scalable compute resources with quicker turnaround times for resource intensive jobs.
Regardless of which resource tier you choose, your data receives the same exceptional level of protection, including a tamper-proof encrypted change log for all of your records.
The user interface wt works on desktop computers, tablets and phones so you can access your data from anywhere with an Internet connection.
Ordium provides tools for Genetic Counselors. It automatically prioritizes interesting and potentially actionable variants that warrant further investigation, so you can skip going over the thousands of irrelevant variants in a sample. It reports the best evidence, from variant read support to automated literature searches, to streamline your workflow and increase throughput.
Base Calling and Alignment
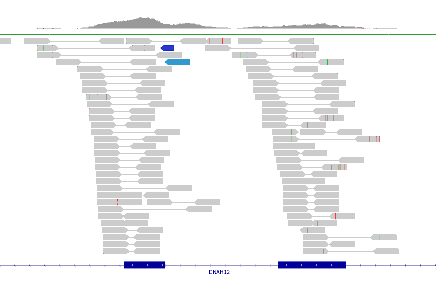
Our primary sequence analysis consists of the initial alignment or assembly of unordered nucleotide fragment data that come from sequence libraries which are captured and output as raw data files on a variety of sequencing devices. We check them for quality, map, sort, mark duplicates, and gather other metrics before we can proceed to call variants.
We've carefully selected the most reliable, industry standard analytic pipelines which have been validated against known reference datasets.

Ordium helps bridge the gap between medicine and biology, using software
Pictured: Trimmed sequence reads with their corresponding base quality scores. Our analyses capture and analyze quality metrics at every step of the process to ensure you are getting the best data from your samples.
Variant Calling
Ordium detects of variations in sequence data with respect to a reference. These variants can be single nucleotide variants (SNVs, or SNPs for polymorphisms), insertions, deletions (indels), or more complex structural variations such as duplications, copy number variations (CNVs), inversions, transversions, gene fusions, or complex structural rearrangements consisting of multiple junctions, generating a variety of RNA isoforms and protein structure.
At this stage we try to predict the functional effect on the RNA or protein product of a variant within a gene or transcript using predictive modeling tools. Ordium achieves this using a number of variant calling pipelines which we developed using validated open source tools that we found to have the best balance of sensitivity at detecting variants, and specificity to minimize false positives.
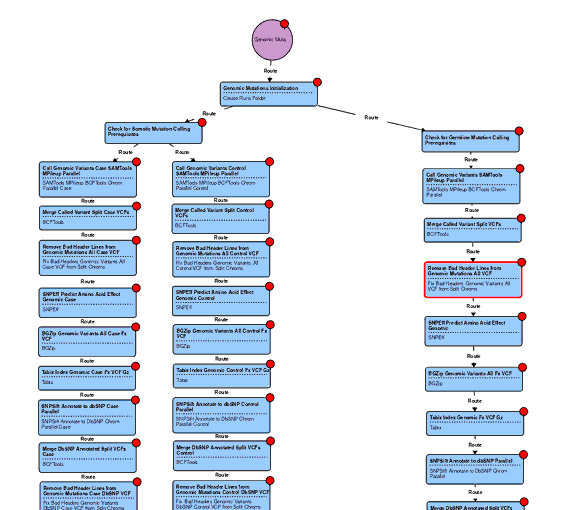
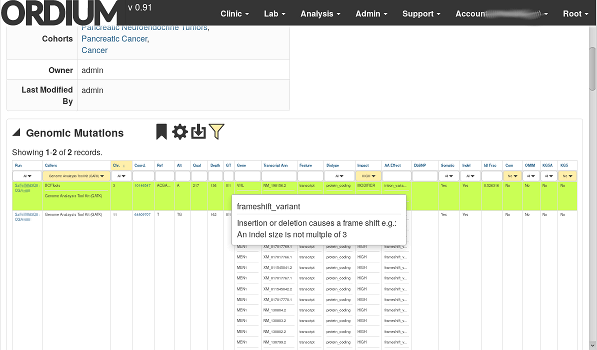
By calling variants in mapped sequence data, then annotating the calls to predict which genes are affected and how, we're able to prioritize a short list of actionable variants in a sample that a clinician or researcher may wish to follow up on.
What Ordium excels at is bringing your interesting variant calls to the top of the list. It's not practical to comb through roughly 30,000 variants in a human exome sample (most of which are irrelevant). We have sensible prioritization defaults, but it's also easy to customize, so you can narrow your search if you're looking for something specific, or broaden it with just a few clicks to explore the data.
If there is ambiguity in a variant call and you need to look at the raw sequence read data, it's right there. Every sequencing technology has limitations, and knowing the limitations of the technology, along with having a good interface to quickly drill down to the raw data improves confidence in your findings.
The best way to learn more about these tools is to Sign up for free. or Contact us to schedule a demo.
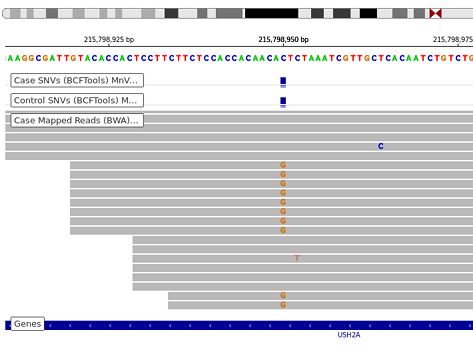
Cohort Analysis
One of the major challenges in working with genomic data is that more than just a few variants will typically be detected in a sample. For example in a human genome, there may be over a million polymorphisms. ("many change" common variations among populations) the vast majority of which are benign. Even in somatic mutation calling from a case/control pair such as tumor vs normal in cancer, there will typcially hundreds or even thousands of somatic mutations called, but very few with a direct causal relationship.
Ordium helps you take advantage of open source knowledge bases to put your called variants in context, whether n=1 or n=1000. It provides tools to perform Genome wide Association Studies (GWAS) on cohorts of samples of virtually any size.
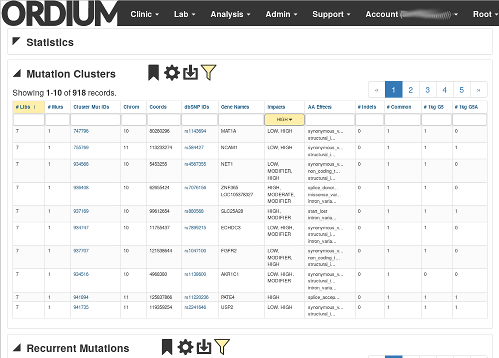
Ordium cohort analysis detects recurrent variants within populations, giving statistical power to a predicted association between a disease or phenotype, and variants which are called. Raw data is also available for download, to support external tools.
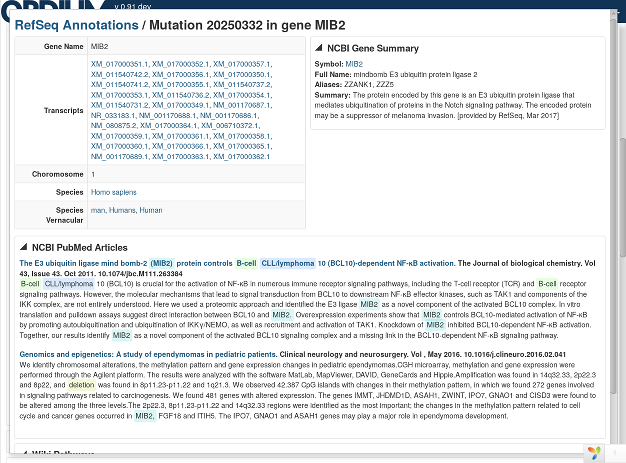
Ordium has the capability to perform quick, automated literature searches based on affected genes and sample and cohort metadata, to dramatically speed up this repetitive manual process of checking if variants have been reported in the literature in association with a disease or phenotype.
A lot of what we've done with has been based on our experience working with researchers and clinicians, knowing where the bottlenecks are and how to streamline workflows to facilitate research and discovery.
Ordium is designed with the flexibility to add new pipelines and support new technologies as they become available. Genomics is a rapidly evolving field, so it's important to stay on top of technology trends. We know there are a lot of talented people out there working in this space, and we want to hear from you if you have suggestions or want to help steer the future development of this project.



